All the examples might not be up to date on the website, please go to our GitLab repository and download latest version of the notebook.
Simple dose computation and optimization on a real CT image
Author : Eliot Peeters
In this example we are going to see how to :
- Import real dicom images and RT struct
- Create a plan
- Compute beamlets
- Optimize a plan with beamlets
- Save a plan and beamlets
- Compute DVH histograms
import numpy as np
import os
from matplotlib import pyplot as plt
#Import the needed opentps.core packages
from opentps.core.data.plan import PlanDesign
from opentps.core.data import DVH
from opentps.core.io import mcsquareIO
from opentps.core.io.scannerReader import readScanner
from opentps.core.processing.doseCalculation.doseCalculationConfig import DoseCalculationConfig
from opentps.core.processing.doseCalculation.mcsquareDoseCalculator import MCsquareDoseCalculator
from opentps.core.io.dataLoader import readData
from opentps.core.data.plan import ObjectivesList
from opentps.core.data.plan import FidObjective
from opentps.core.io.serializedObjectIO import saveBeamlets, saveRTPlan, loadBeamlets, loadRTPlan
In the next cell we configure the CT scan model used for the dose calculation and the bdl model. The ones used in this example are the default configuration of openTPS wich may lead to some imprecision.
ctCalibration = readScanner(DoseCalculationConfig().scannerFolder)
bdl = mcsquareIO.readBDL(DoseCalculationConfig().bdlFile)
Data importation
The dataset used in this example comes from the Proknow website, 2018 TROG Plan Study: SRS Brain. The readData functions automatically import the subfolders and detects the type of data (CT or RT_struct).
ctImagePath = "Path_to_\ProKnows_2018_TROG_Plan_Study_SRS_Brain" #The folder is initially named 'data'
data = readData(ctImagePath)
rt_struct = data[0]
ct = data[1]
rt_struct.print_ROINames()
RT Struct UID: 2.16.840.1.114362.1.6.6.9.161209.11109530676.470491132.964.204
[0] GTV1-20Gy
[1] GTV2-20Gy
[2] GTV3-20Gy
[3] GTV4-20Gy
[4] Eye_L
[5] Eye_R
[6] Lens_L
[7] Lens_R
[8] OpticNerve_L
[9] OpticNerve_R
[10] Optic Chiasm
[11] Hippocampus_L
[12] Hippocampus_R
[13] Brainstem
[14] Brain
[15] BODY
[16] Bones
[17] Normal Brain
[18] GTV5-20Gy
[19] GTV-Total
For the purpose of the demonstration we are going to use only 3 different ROI. Note that it is important to specify the CT origin,gridSize and spacing to the getBinaryMask function in order to have a correct binary mask.
target_name = "GTV4-20Gy"
target = rt_struct.getContourByName(target_name).getBinaryMask(origin=ct.origin,gridSize=ct.gridSize,spacing=ct.spacing)
OAR_brain = rt_struct.getContourByName("Brain").getBinaryMask(origin=ct.origin,gridSize=ct.gridSize,spacing=ct.spacing)
OAR_brainstem = rt_struct.getContourByName("Brainstem").getBinaryMask(origin=ct.origin,gridSize=ct.gridSize,spacing=ct.spacing)
OAR_optic_chiasm = rt_struct.getContourByName("Optic Chiasm").getBinaryMask(origin=ct.origin,gridSize=ct.gridSize,spacing=ct.spacing)
For further plots we can extract the indexes of the centerOfMass of the tumor.
COM_coord = target.centerOfMass
COM_index = target.getVoxelIndexFromPosition(COM_coord)
Z_COORD = COM_index[2]
MCsquare configuration
We now initialize a MCsquareDoseCalculator and provide the beamModel and ctCalibration imported above.
# Configure MCsquare
mc2 = MCsquareDoseCalculator()
mc2.beamModel = bdl
mc2.ctCalibration = ctCalibration
Plan design
In the next section we create a planDesign object with 3 beams (of no medical relevance, we just use them for demonstration). There are multiple parameters which can affect computation time :
targetMargin: a higher margin will increase the time used to dilate the maskspotSpacing: a lower spot spacing will result in more beamlets therefore longer beamlets calculation timelayerSpacing: a lower layer spacing will result in more beamlets therefore longer beamlets calculation time
# Design plan
beamNames = ["Beam1","Beam2","Beam3"]
gantryAngles = [0.,45.,315.]
couchAngles = [0.,0.,0.]
# Generate new plan
planDesign = PlanDesign()
planDesign.ct = ct
planDesign.gantryAngles = gantryAngles
planDesign.targetMask = target
planDesign.beamNames = beamNames
planDesign.couchAngles = couchAngles
planDesign.calibration = ctCalibration
planDesign.spotSpacing = 5.0
planDesign.layerSpacing = 5.0
planDesign.targetMargin = 5.0
plan = planDesign.buildPlan() # Spot placement
plan.PlanName = "NewPlan"
08/08/2023 09:33:47 AM - opentps.core.data.plan._planDesign - INFO - Building plan ...
08/08/2023 09:33:49 AM - opentps.core.processing.planOptimization.planInitializer - INFO - Target is dilated using a margin of 5.0 mm. This process might take some time.
08/08/2023 09:33:49 AM - opentps.core.processing.imageProcessing.roiMasksProcessing - INFO - Using SITK to dilate mask.
08/08/2023 09:34:32 AM - opentps.core.data.plan._planDesign - INFO - New plan created in 44.99794363975525 sec
08/08/2023 09:34:32 AM - opentps.core.data.plan._planDesign - INFO - Number of spots: 609
Beamlets computation and initial dose computation
In the next section we compute the beamlets (this is the most computer-intensive part). We have set the numbers of protons to 5e4.
mc2.nbPrimaries = 5e4
beamlets = mc2.computeBeamlets(ct, plan)
plan.planDesign.beamlets = beamlets
After the beamlets computation we can save the plan and the beamlets to reuse them in the future.
WARNING : the
saveRTPlanfunction automatically remove the beamlets from the memory, if you want to save the beamlets, you have to call thesaveBeamletsfunction before. Those files can be heavy !
Afterward you can load the plan and the beamlets via the loadRTPlan and loadBeamlets functions.
#Output path
output_path = 'Output'
if not os.path.exists(output_path):
os.makedirs(output_path)
# Save the plan and the beamlets
saveBeamlets(beamlets, os.path.join(output_path, "SimpleRealDoseComputationOptimization_beamlets.blm"))
saveRTPlan(plan, os.path.join(output_path,"SimpleRealDoseComputationOptimization_plan.tps"))
plan = loadRTPlan(os.path.join(output_path,"SimpleRealDoseComputationOptimization_plan.tps"))
plan.planDesign.beamlets = loadBeamlets(os.path.join(output_path,"SimpleRealDoseComputationOptimization_beamlets.blm"))
Note that in the next cell we have augmented the number of protons for the dose computation (computeDose) to have a more accurate dose. This dose is computed with all the weights of the beamlets set to 1.
mc2.nbPrimaries = 1e7
dose_before_opti = mc2.computeDose(ct,plan)
Plan optimization
We will now optimize the plan with and without OAR to compare the differences. We first create an ObjectivesList and then add objectives via the addFidObjective which can be either DMIN, DMAX or DMEAN. Note that you can also create other objectives and implement them via the addExoticObjective fucntion.
plan.planDesign.objectives = ObjectivesList() #create a new objective set
plan.planDesign.objectives.setTarget(target.name, 20.0) #setting a target of 20 Gy for the target
plan.planDesign.objectives.fidObjList = []
plan.planDesign.objectives.addFidObjective(target, FidObjective.Metrics.DMAX, 19.5, 1.0)
plan.planDesign.objectives.addFidObjective(target, FidObjective.Metrics.DMIN, 20.5, 1.0)
plan.planDesign.defineTargetMaskAndPrescription()
We will use the Scipy-LBFGS as solver for this example but other are also implemented such as :
- Scipy-BFGS
- Scipy-LBFGS
- Gradient
- BFGS
- LBFGS
- FISTA
- LP
Feel also free to specify a maxit to the IMPTPlanOptimizer object to speed up the program.
from opentps.core.processing.planOptimization.planOptimization import IMPTPlanOptimizer
solver = IMPTPlanOptimizer(method='Scipy-LBFGS',plan=plan)
w, doseImage, ps = solver.optimize()
plan.spotMUs = np.square(w).astype(np.float32)
We can now recompute the dose.
doseImage_opti = mc2.computeDose(ct,plan)
We here reload the plan in order to reset all weights and the filtering (after optimization, spots that are bellow the solver.thresholdSpotRemoval and corresponding weights are removed).
plan = loadRTPlan(os.path.join(output_path,"SimpleRealDoseComputationOptimization_plan.tps"))
plan.planDesign.beamlets = loadBeamlets(os.path.join(output_path,"SimpleRealDoseComputationOptimization_beamlets.blm"))
plan.planDesign.objectives = ObjectivesList() #create a new objective set
plan.planDesign.objectives.setTarget(target.name, 20.0) #setting a target of 20 Gy for the target
plan.planDesign.objectives.fidObjList = []
plan.planDesign.objectives.addFidObjective(target, FidObjective.Metrics.DMAX, 20.5, 1.0)
plan.planDesign.objectives.addFidObjective(target, FidObjective.Metrics.DMIN, 19.5, 1.0)
plan.planDesign.objectives.addFidObjective(OAR_brain, FidObjective.Metrics.DMAX, 8.0, 1.0)
plan.planDesign.objectives.addFidObjective(OAR_brainstem, FidObjective.Metrics.DMAX, 5.0, 1.0)
plan.planDesign.objectives.addFidObjective(OAR_optic_chiasm, FidObjective.Metrics.DMAX, 2.0, 1.0)
plan.planDesign.defineTargetMaskAndPrescription()
solver = IMPTPlanOptimizer(method='Scipy-LBFGS',plan=plan)
doseImage, ps = solver.optimize()
doseImage_opti_OAR = mc2.computeDose(ct,plan)
DVH histograms
We can create simple DVH plots with the DVH objects. Take a look at the class properties to find the D95, …
target_DVH = DVH(target,doseImage_opti_OAR)
target_DVH_No_OAR = DVH(target,doseImage_opti)
brain_DVH = DVH(OAR_brain,doseImage_opti_OAR)
brainstem_DVH = DVH(OAR_brainstem,doseImage_opti_OAR)
optic_chiasm_DVH = DVH(OAR_optic_chiasm,doseImage_opti_OAR)
Final plots
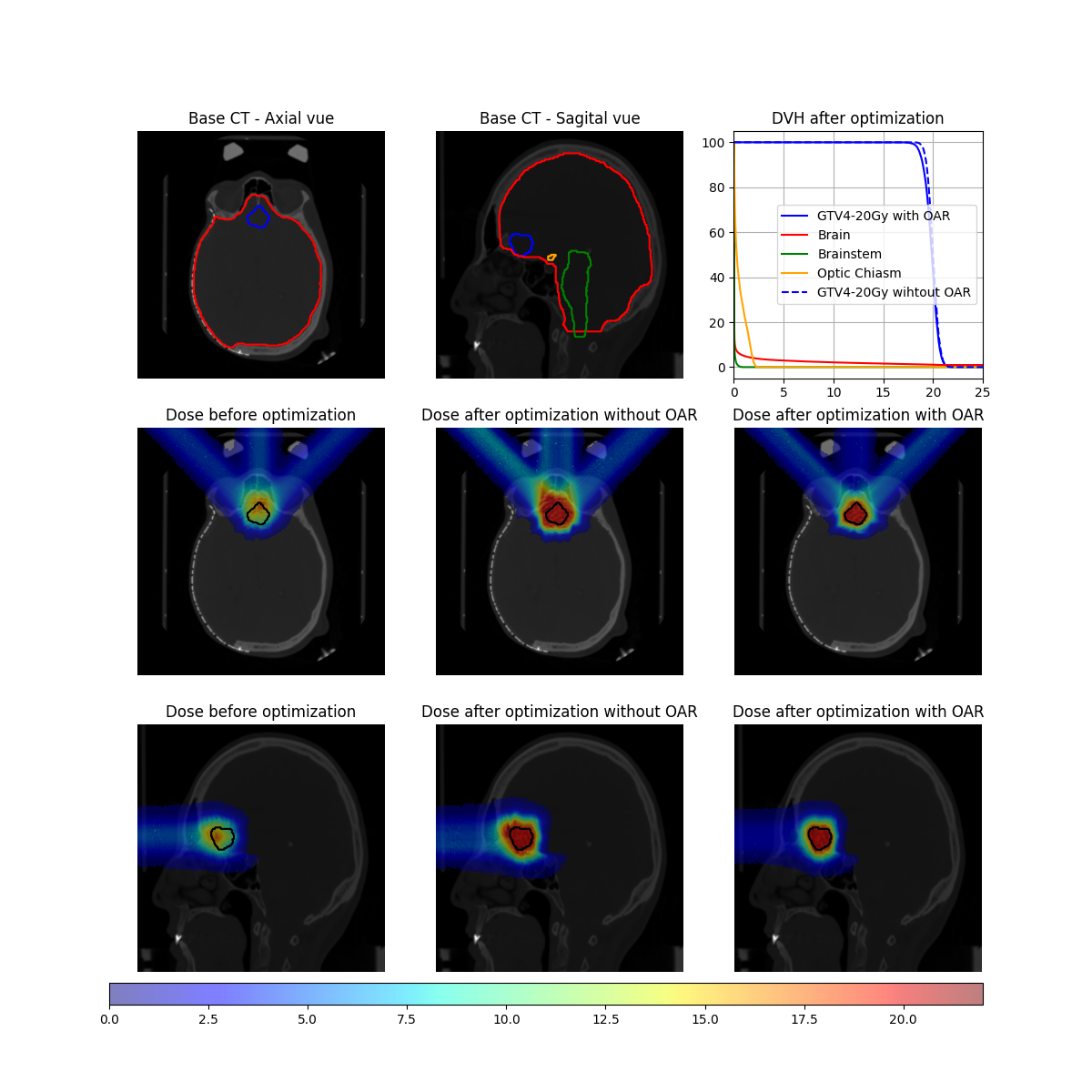
Now that the different optimizations are done, we can display the results.
from skimage.transform import resize
image_ct_axial = ct.imageArray[:,:,Z_COORD].transpose(1,0)
image_target_axial = target.imageArray[:,:,Z_COORD].transpose(1,0)
image_brain_axial = OAR_brain.imageArray[:,:,Z_COORD].transpose(1,0)
image_brainstem_axial = OAR_brainstem.imageArray[:,:,Z_COORD].transpose(1,0)
image_optic_chiasm_axial = OAR_optic_chiasm.imageArray[:,:,Z_COORD].transpose(1,0)
image_dose_before_opti_axial = dose_before_opti.imageArray[:,:,Z_COORD].transpose(1,0)
image_dose_opti_axial = doseImage_opti.imageArray[:,:,Z_COORD].transpose(1,0)
image_dose_opti_OAR_axial = doseImage_opti_OAR.imageArray[:,:,Z_COORD].transpose(1,0)
image_ct_sagital = resize(np.rot90(ct.imageArray[COM_index[0],:,:]),(ct.gridSize[:-1]))
image_target_sagital = resize(np.rot90(target.imageArray[COM_index[0],:,:]),(ct.gridSize[:-1]))
image_brain_sagital = resize(np.rot90(OAR_brain.imageArray[COM_index[0],:,:]),(ct.gridSize[:-1]))
image_brainstem_sagital = resize(np.rot90(OAR_brainstem.imageArray[COM_index[0],:,:]),(ct.gridSize[:-1]))
image_optic_chiasm_sagital = resize(np.rot90(OAR_optic_chiasm.imageArray[COM_index[0],:,:]),(ct.gridSize[:-1]))
image_dose_before_opti_sagital = resize(np.rot90(dose_before_opti.imageArray[COM_index[0],:,:]),(ct.gridSize[:-1]))
image_dose_opti_sagital = resize(np.rot90(doseImage_opti.imageArray[COM_index[0],:,:]),(ct.gridSize[:-1]))
image_dose_opti_OAR_sagital = resize(np.rot90(doseImage_opti_OAR.imageArray[COM_index[0],:,:]),(ct.gridSize[:-1]))
min_dose_val = 0.1
image_dose_before_opti_axial[image_dose_before_opti_axial <= min_dose_val] = np.nan
image_dose_opti_axial[image_dose_opti_axial <= min_dose_val] = np.nan
image_dose_opti_OAR_axial[image_dose_opti_OAR_axial <= min_dose_val] = np.nan
image_dose_before_opti_sagital[image_dose_before_opti_sagital <= min_dose_val] = np.nan
image_dose_opti_sagital[image_dose_opti_sagital <= min_dose_val] = np.nan
image_dose_opti_OAR_sagital[image_dose_opti_OAR_sagital <= min_dose_val] = np.nan
vmin=0
vmax=22
fig, ax = plt.subplots(3,3,figsize=(12,12))
ax[0,0].imshow(image_ct_axial,cmap="gray")
ax[0,0].contour(image_target_axial,colors="blue")
ax[0,0].contour(image_brain_axial,colors="red")
ax[0,0].contour(image_brainstem_axial,colors="green")
ax[0,0].contour(image_optic_chiasm_axial,colors="orange")
ax[0,0].axis("off")
ax[0,0].set_title("Base CT - Axial vue")
ax[0,1].imshow(image_ct_sagital,cmap="gray")
ax[0,1].contour(image_target_sagital,colors="blue")
ax[0,1].contour(image_brain_sagital,colors="red")
ax[0,1].contour(image_brainstem_sagital,colors="green")
ax[0,1].contour(image_optic_chiasm_sagital,colors="orange")
ax[0,1].axis("off")
ax[0,1].set_title("Base CT - Sagital vue")
ax[0,2].plot(target_DVH.histogram[0],target_DVH.histogram[1],label=target_DVH.name + " with OAR",color="blue")
ax[0,2].plot(brain_DVH.histogram[0],brain_DVH.histogram[1],label=brain_DVH.name, color="red")
ax[0,2].plot(brainstem_DVH.histogram[0],brainstem_DVH.histogram[1],label=brainstem_DVH.name, color="green")
ax[0,2].plot(optic_chiasm_DVH.histogram[0],optic_chiasm_DVH.histogram[1],label=optic_chiasm_DVH.name, color="orange")
ax[0,2].plot(target_DVH_No_OAR.histogram[0],target_DVH_No_OAR.histogram[1],label=target_DVH.name + " wihtout OAR",color="blue",linestyle="dashed")
ax[0,2].set_xlim(0,25)
ax[0,2].grid(True)
ax[0,2].legend()
ax[0,2].set_title("DVH after optimization")
ax[1,0].imshow(image_ct_axial,cmap="gray")
dose_bar_ref = ax[1,0].imshow(image_dose_before_opti_axial,cmap="jet",alpha=.5,vmin=vmin,vmax=vmax)
ax[1,0].contour(image_target_axial,colors="black")
ax[1,0].set_title("Dose before optimization")
ax[1,0].axis("off")
ax[1,1].imshow(image_ct_axial,cmap="gray")
ax[1,1].imshow(image_dose_opti_axial,cmap="jet",alpha=.5,vmin=vmin,vmax=vmax)
ax[1,1].contour(image_target_axial,colors="black")
ax[1,1].set_title("Dose after optimization without OAR")
ax[1,1].axis("off")
ax[1,2].imshow(image_ct_axial,cmap="gray")
ax[1,2].imshow(image_dose_opti_OAR_axial,cmap="jet",alpha=.5,vmin=vmin,vmax=vmax)
ax[1,2].contour(image_target_axial,colors="black")
ax[1,2].set_title("Dose after optimization with OAR")
ax[1,2].axis("off")
ax[2,0].imshow(image_ct_sagital,cmap="gray")
ax[2,0].imshow(image_dose_before_opti_sagital,cmap="jet",alpha=.5,vmin=vmin,vmax=vmax)
ax[2,0].contour(image_target_sagital,colors="black")
ax[2,0].set_title("Dose before optimization")
ax[2,0].axis("off")
ax[2,1].imshow(image_ct_sagital,cmap="gray")
ax[2,1].imshow(image_dose_opti_sagital,cmap="jet",alpha=.5,vmin=vmin,vmax=vmax)
ax[2,1].contour(image_target_sagital,colors="black")
ax[2,1].set_title("Dose after optimization without OAR")
ax[2,1].axis("off")
ax[2,2].imshow(image_ct_sagital,cmap="gray")
ax[2,2].imshow(image_dose_opti_OAR_sagital,cmap="jet",alpha=.5,vmin=vmin,vmax=vmax)
ax[2,2].contour(image_target_sagital,colors="black")
ax[2,2].set_title("Dose after optimization with OAR")
ax[2,2].axis("off")
cb_ax = fig.add_axes([0.1, 0.08, 0.8, 0.02])
fig.colorbar(dose_bar_ref,cax=cb_ax,location="bottom")
plt.savefig(os.path.join(output_path, "SimpleRealDoseComputationOptimization_output.png"))
plt.close()
Download this notebook via our GitLab repository